FRONTIERS The Promise and Perils of Big Data
“Big data,” says Salk Research Professor Margarita Behrens, “is like looking at the night sky and seeing a beautiful, bright star, and then looking through a telescope and having that star get lost against the backdrop of so many others. Initially you can lose your bearings.”
The concept of “big data” refers to the idea that vast quantities of data too unwieldy for human calculation can be manipulated and analyzed for meaningful patterns using computational algorithms. The idea has been around for decades, but only recently has a sufficient amount of data generation coincided with enough computing power to truly begin to realize the potential—and the challenges—of big data.
Billions of Bases of DNA, and Beyond
In 2013, Behrens, along with Salk professors Joseph Ecker and Terrence Sejnowski, published a groundbreaking paper in the journal Science and, as she jokes, “opened Pandora’s box.”
The paper, for which Behrens and Ecker were the senior authors, provided the first comprehensive maps of epigenomic changes in the brain, known as DNA methylation. These chemical tags on a cell’s DNA act as an extra layer of information, overlying the genome and controlling when genes are turned on or off. Behrens refers to the study results as Pandora’s box because the “data was so rich.”
The researchers set out with what initially seemed like a fairly straightforward goal: cataloging methylation patterns in mouse and human brain samples at different stages of brain development. But, to everyone’s surprise, the data revealed a previously unknown level of complexity: the methylation patterns turned out to be specific to whether brain cells were neurons or glia (neurological support cells), which underwent extensive reconfigurations during different developmental stages. The discovery that methylation could be this specific to subtypes of cells opened up a whole new field of identifying and characterizing brain cells based on an ever-expanding set of cellular features including DNA sequence, DNA packaging, RNA sequence and others.
The work was surprising not only for what it revealed about the complexity and dynamism of brain circuitry, but also for what it showed about the power of big data to open up entire new universes of discovery.

It would take the human brain 190,000 YEARS to read the data from just one 48-hour DNA-sequencing run from a high-throughput sequencer
Ecker, who directs Salk’s Genomic Analysis Laboratory, is keenly aware of how much data scientists can now compute, not just for the epigenome but also to better understand new facets of the genome. Outside Ecker’s office is a humming instrument called the NovaSeq 6,000, which is busily sequencing DNA. In 48 hours, it categorizes six terabases of DNA (that is 6 followed by 12 zeros—6,000,000,000,000). That’s 6 trillion bases (letters), an amount that is far too vast for the human mind to comprehend. If it took you one second to read each letter, it would take 190,000 years to read all the letters from just one 48-hour DNA-sequencing run.
And this is 6,000 times greater than the first next-generation sequencer that Ecker had purchased only about a dozen years ago, in 2007. So, large computing power is essential for “reading” all of this information.
“We are now to the point where we’re looking at the DNA sequences of individual cells from millions of cells in the brain,” says Ecker, who is also a Howard Hughes Medical Institute investigator.
Ecker and collaborators, both at Salk and elsewhere, are trying to capture different kinds of genetic data—what the genes are, how they’re being regulated, how the chromosomes are folding and how the genes are being expressed—all from the same cell, to develop a comprehensive understanding of individual brain cells. By developing such detailed blueprints of healthy cells, scientists will be better able to pinpoint what goes wrong in a variety of diseases, such as cancer and developmental or neurological disorders.
Ecker, Salk-Helmsley Fellow Jesse Dixon and collaborators recently published a pair of papers describing big data computation tools and techniques they have developed, making headway in this area. The first paper, published in Proceedings of the National Academy of Sciences, details an algorithm that lets researchers identify cells based on the shape of their chromosomes. The second paper, published in Nature Methods, shows how the team combined two different analysis techniques into one method, enabling the team to identify gene regulatory elements in distinct cell types. Because the majority of the human genome is made up of regulatory DNA—stretches of DNA that don’t themselves encode proteins but that help control whether, and when, genes are expressed in any given cell—insights into the regulatory aspects of the genetic code have the potential to greatly further our understanding of health and disease. (See this issue’s “Discoveries” for details.)
Computational analysis indicated that the brain’s memory capacity is 10 times greater than had been thought.
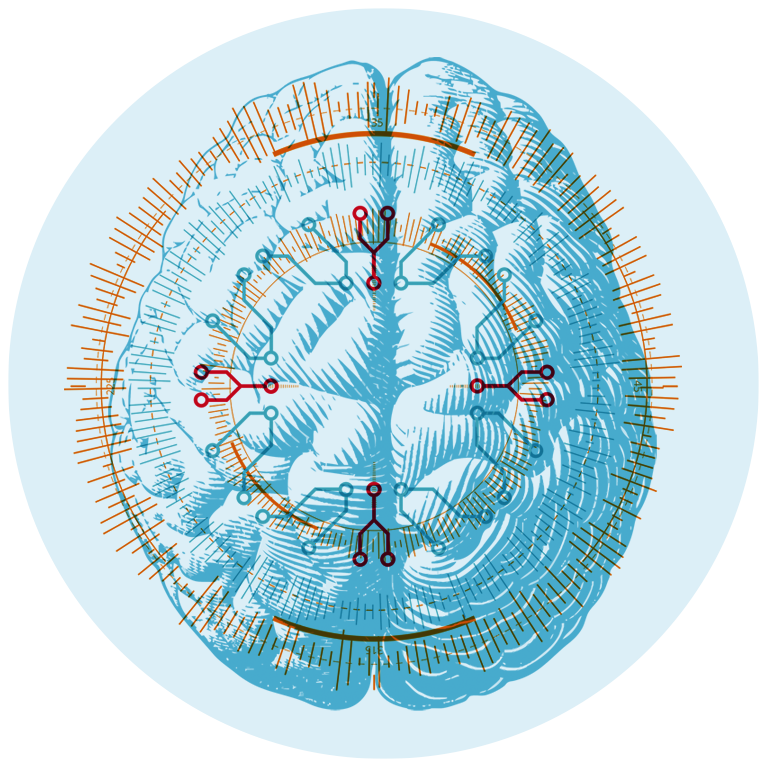
This Is Your Brain On Big Data
Big data is a direct result of progress in computing and artificial intelligence (AI), the effort to have machines mimic the intelligence of the human brain. Advances in the field of neuroscience, therefore, have greatly informed AI. Sejnowski, head of the Computational Neurobiology Laboratory and holder of the Francis Crick Chair, has been at the leading edge of the AI movement since the 1980s.
“The brain has been in the big data business for a lot longer than we have,” Sejnowski says. Indeed, the brain has been the epitome of big data crunching—somehow it is able to process and make sense of enormous amounts of data. This data deluge can also overwhelm a brain, which must decide what data to save and what data to ignore. Thus, brains have become an inspiration for new ways to develop AI and other tools. As Sejnowski writes in his 2018 book, The Deep Learning Revolution: Artificial Intelligence Meets Human Intelligence, “The recent progress in artificial intelligence was made by reverse engineering brains. Learning algorithms in deep learning networks are inspired by the way that neurons communicate with one another and are modified by experience.”
In 2018, as described in the journal Nature, Sejnowski and collaborators at UC San Diego trained robotic gliders to soar like birds using an AI approach called reinforcement learning, inspired by how animals learn to adjust their behavior based on the outcome of their actions to achieve goals. The researchers designed field experiments in which the gliders learned on their own how to soar upwards in thermals like birds, results that could help improve the efficiency of unmanned aerial vehicles.
Apart from his expertise on various types of artificial intelligence, Sejnowski has used advanced microscopy and computational algorithms to make profound discoveries about memory, including finding that the brain’s memory capacity is 10 times greater than had been thought, and that following stimulation, the sizes of synapses—the junctions between neurons where memories are stored—can expand and contract as needed, increasing the range of sizes like an expandable file folder. These findings were detailed in eLife in 2016 and in Proceedings of the National Academy of Sciences in 2018, respectively.
And breakthroughs related to data are only speeding up. As Sejnowski writes in his book, “Data are the new oil. Learning algorithms are refineries that extract information from raw data; information can be used to create knowledge; knowledge leads to understanding; and understanding leads to wisdom.”
Professor Tatyana Sharpee, like Sejnowski, is a member of the Computational Neurobiology Laboratory, where she focuses on understanding how the brain’s billions of neurons exchange information to process sensory information, such as sights, sounds and smells. To decipher vast amounts of experimental data, she applies mathematical strategies, including statistics and probability models.
In one example, Sharpee’s lab yielded a statistical way to understand odor data, which the team was then able to map to discover regions of odor combinations humans find most pleasurable—work that could open new avenues for food scientists, allowing them to engineer smells and tastes. In another discovery, Her lab also used a statistical method to discover that neurons in a region of the brain called V2, which is part of the visual system, were responding to combinations of edges in scenes from nature, such as the outlines of leaves and the texture of bark. The work might improve object-recognition algorithms for self-driving cars or other robotic devices.
“It seems that every time we add elements of computation that are found in the brain to computer-vision algorithms, their performance improves,” Sharpee says. “The challenge is to have the courage to follow the directions where data and experiments lead us rather than settling on approaches that yield immediate results.”
The promise of big data, then, is that meaningful patterns are waiting to be discovered within sets of signals; the challenge is that not all patterns will be meaningful. And in the worst cases, results from machine learning may amplify the biases brought by human interpretation and judgement.
“We are now to the point where we’re looking at the DNA sequences of individual cells from millions of cells in the brain.” – Joseph Ecker
Using Computation To Inform Cancer Research
Cancer biologist Edward Stites is particularly sensitive to both the benefits and the detriments inherent in big data approaches. In his field, time spent chasing down false leads can mean delaying the pursuit of promising ones.
“As a physician who previously trained in mathematics, I completely appreciate the importance of bringing more mathematics, more computation and more data analysis to problems of medical importance,” says Stites, an assistant professor in the Integrative Biology Laboratory. “But as someone who has trained in both of these areas, I also think it is critical that these efforts take careful consideration of both the underlying mathematics of utilized methods and the biology that is being studied.”
Stites works on problems in cancer biology—a field where the multifaceted nature of the disease complicates progress by traditional methods. However, his lab is using mathematical and computational methods to extrapolate from available data to generate completely new ideas for solutions to those problems. In a paper published in Science Signaling in September 2019, Stites and first author Thomas McFall, a Salk postdoctoral fellow, describe how, using a combination of computational and experimental methods, they discovered the mechanism for how the colorectal cancer drug cetuximab works. (See this issue’s “Discoveries” section for details.) Knowing how a drug works helps doctors feel more confident in prescribing it, which could benefit upwards of 10,000 colorectal cancer patients per year, according to the researchers.
Stites’ team first used computational models to simulate complex reactions and tease out differences between healthy genes and mutant colorectal cancer genes based purely on biochemical information, yet with the ability to explain what was seen in a clinical trial. Their mathematics told them where to look in their laboratory tests to identify the molecular mechanism by which colorectal cancer patients with a specific gene mutation (KRAS G13D) responded to cetuximab. The researchers then replicated their findings across three genetically distinct cell lines to demonstrate the reliability of the results.
The work is an example of how, in the best cases, big data is a tool that enhances researchers’ approaches. Computers may find patterns, but you need humans to understand their meaning.
“Einstein from noise” is a cautionary example from cryo-EM.
An algorithm trained using images of Albert Einstein along with 1,000 images of pure white noise resulted in a pattern of Einstein’s face.

Separating the Signal From the Noise
Assistant Professor Dmitry Lyumkis says that in his field of extremely high-resolution microscopy known as cryo-EM, there’s a well-known cautionary example called “Einstein from noise.” Just as static on the radio can make it hard to hear music clearly, scientific “noise” refers to an unwanted signal that interferes with information of interest. So, in the cryo-EM example, researchers trained an algorithm using images of Albert Einstein. When they then ran 1,000 images of pure white noise through the algorithm, the pattern it found was … Einstein’s face. The pattern was not really there, but the algorithm found it based on how it had been trained.
“It was a case of garbage in, garbage out,” says Lyumkis, echoing Stites’ notion that researchers need to pay careful attention to what their training algorithms are learning, because if the algorithms are not gleaning the right lessons, any results they offer could at best be useless and at worst lead research astray.
As a structural biologist, Lyumkis uses cryo-EM to directly visualize large molecular assemblies isolated from cells under near-native conditions. Because molecular structure is so intimately tied to function in biology, by better understanding the architecture and atomic-level details of large molecular assemblies, scientists may gain clarity on various types of dysfunction that lead to disease, such as HIV. In 2017, in work published in Science, Lyumkis’ lab solved the atomic structure of a key protein machine called an “intasome,” which allows HIV to integrate into human host DNA and replicate in the body. HIV intasome structures have eluded researchers for decades. The findings yield atomic-level clues that are now informing the development of new drugs.
Lyumkis’ field has particularly benefited from advances in computing power. Cryo-EM data sets can comprise hundreds of thousands of individual images that have to be computationally analyzed in order to create an accurate three-dimensional reconstruction of a biological sample.
Lyumkis says that at the turn of the millennium, the field was struggling to resolve molecular assemblies to better than 1 nanometer resolution (one nanometer is 100,000 times smaller than the diameter of a hair). By 2008, cryo-EM researchers were able to achieve three times better resolution.
“One way to think about it is that, between the years of 2000 and 2010, microscopes and samples didn’t change very much,” Lyumkis says. “But what changed is the computing power and algorithms and how much data we can crunch. Improvements in computing power and our ability to make sense of increasingly more complex datasets at higher resolution are continuing to extend the boundaries of the types of questions we can ask.”
The Big Picture
Salk Vice President, Chief Science Officer and Professor Martin Hetzer is keen to harness the power of big data by incorporating computational approaches that enhance the experimental approaches of bench science for which the Institute is renowned.
In December 2018, Hetzer and a collaborative team at Salk published a paper in Genome Biology describing how they used machine learning to analyze skin cells from the very young to the very old to find molecular signatures that can be predictive of age. Using custom algorithms to sort the data, the team found certain biomarkers indicating aging, and were able to predict a person’s age with less than four years of error on average.
“The next big trend in science is about understanding massive amounts of data and will take new kinds of collaborations to do so,” Hetzer says.
The process is still ramping up, and scientists need to make sure plenty of checks and balances exist to verify the data and results along the way. But, in the end, Behrens’ Greek myth analogy of big data may be apt, because after problems flew out of Pandora’s box, what was left was hope. And although scientists are often generating more data than they and computers can analyze, they are optimistic that the analysis will eventually catch up, and big data will help lead to answers for some of biology’s most urgent questions.
Support a legacy where cures begin.
Featured Stories
 The Promise and Perils of Big DataSalk scientists are unlocking the power of "big data" to make unprecedented discoveries in neuroscience, cancer and other areas.
The Promise and Perils of Big DataSalk scientists are unlocking the power of "big data" to make unprecedented discoveries in neuroscience, cancer and other areas.
 Joseph Noel – Learning from LagoonsInside Salk sat down with Noel to learn about his path to becoming a scientist and about his current work researching coastal wetlands to help combat climate change.
Joseph Noel – Learning from LagoonsInside Salk sat down with Noel to learn about his path to becoming a scientist and about his current work researching coastal wetlands to help combat climate change.
 Krishna Vadodaria – Uncovering the mysteries of depressionStaff Scientist Krishna Vadodaria's research on human neurons is helping to uncover the biological basis for psychiatric disorders and why some depressed patients do not respond to SSRIs.
Krishna Vadodaria – Uncovering the mysteries of depressionStaff Scientist Krishna Vadodaria's research on human neurons is helping to uncover the biological basis for psychiatric disorders and why some depressed patients do not respond to SSRIs.